Subgroup Discovery
Title : Stratification of psychiatric diseases into homogeneous subgroups
Project coordinator : Edouard Duchesnay
Participants : Pietro Gori and Robin Louiset
Institutions : Télécom Paris and NeuroSpin (CEA)
Funding: PhD CEA funding (110k)
Period : 2020-2024
Context and Challenges - In the past decades, unsupervised and self-supervised learning techniques have proven to be particularly effective at identifying relevant patterns and factors of variation within a dataset. Combined with powerful Neural Networks (NNs), these methods can produce semantically rich representations. Notably, unsupervised Deep Clustering (DC) methods seek to produce a suitable representation space for identifying homogeneous latent clusters based on the general variability of the entire dataset (i.e., imaging patterns common to all samples). With a different perspective, Subgroup Discovery (SD) in medical applications aims at identifying relevant latent subtypes/subgroups that arise from the pathological variability of the diseased population and not from the irrelevant common variability that may exist in both healthy subjects (i.e., controls) and diseased patients.
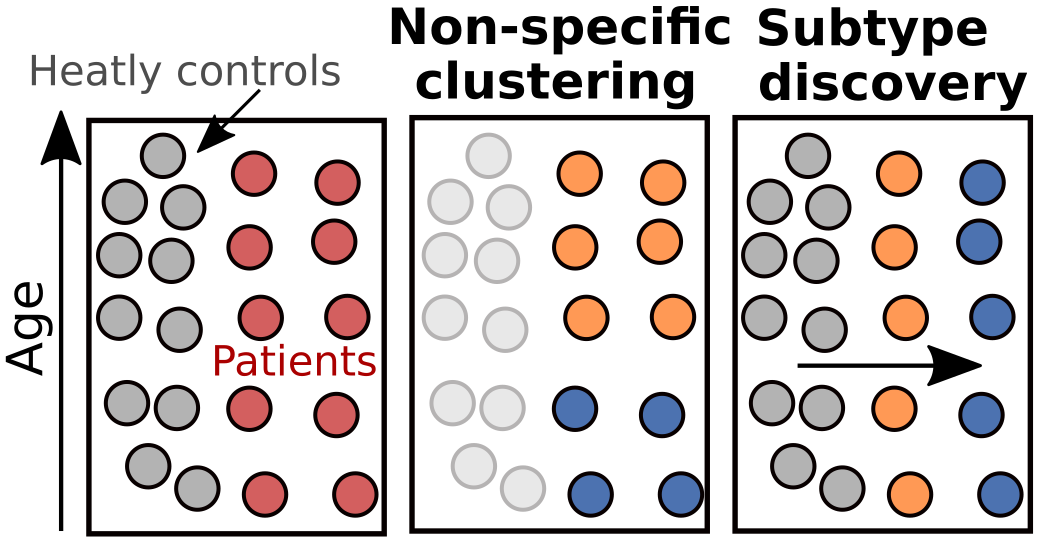
For instance, in clinical research, it is essential to identify subtypes of patients with a given disorder (red dots in the figure above). However, the general variability that stems from age or sex, and which is observed in both healthy controls (grey dots) and patients will probably drive the clustering of patients to a non-specific solution (2nd plot). Instead, subtypes should be defined only by the modes of variation (horizontal arrow) specific to the pathology, thus discarding nonspecific variability and emphasizing more disease-related differences (3rd plot).
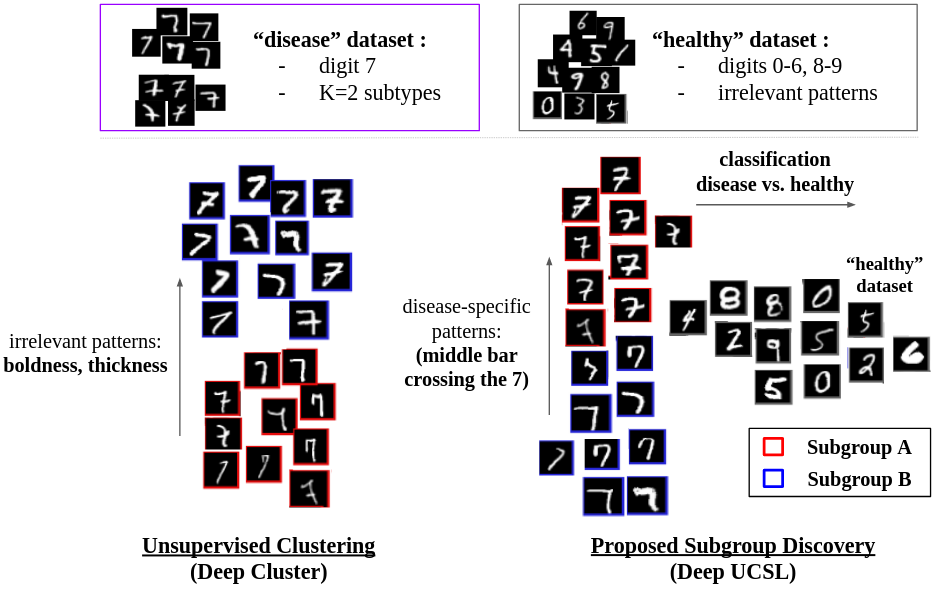
In the figure above, we use an intuitive toy example based on the MNIST dataset to better clarify the differences between Deep Clustering and Subtype Discovery. We consider the digit ”7” as the pathological group and all the other digits as the healthy group. Results show how Deep Cluster’s subgroups of the digit “7” are only defined by the most predominant characteristics (i.e.: boldness of the digit) common to all digits. Instead, a Subtype Discovery method, such as the proposed UCSL or Deep UCSL, disregards these common characteristics and uses only the specific patterns of the digit “7” (i.e., the presence of the crossing middle bar) to define the subgroups.
Goal: Propose new machine-learning methods to better stratify highly-heterogeneous mental disorders using anatomical MRI data. This should help human experts discover or validate subgroups, while relying on reproducible, data-driven, and objective imaging patterns.
Contributions By contrasting controls with patients, we proposed two new methods to identify subgroups that arise solely from the pathological variability specific to the disease, while disregarding the common variability shared with the controls. We named these methods UCSL (Louiset et al., 2021), which is based on linear Machine Learning techniques, and Deep UCSL (Louiset et al., 2024), which utilizes Deep Learning instead. The objectives of both methods are:
a) to accurately identify the pathological subgroups,
b) to ensure that healthy samples are not assigned to a pathological subgroup, and
c) to effectively distinguish each subgroup from the healthy class.
References
2024
- Under ReviewAutomatic Discovery of Disease Subgroups by Contrasting with Healthy Controls.Under Review, 2024