Brain white matter tractogram analysis
Title : Neural Meta Tracts: parsimonious multi-resolution representations for modeling, visualizing and statistically analyzing brain tractograms
Project coordinator : Pietro Gori
Participants : Jean-Marc Thiery, Damien Rohmer, Isabelle Bloch, Tamy Boubekeur, Marie-Paule Cani and Johan Pallud
Institutions : Télécom Paris, Ecole Polytechnique and St. Anne hospital
Funding: DigiCosme Emergence Project (240 k€)
Period : 2017-2020

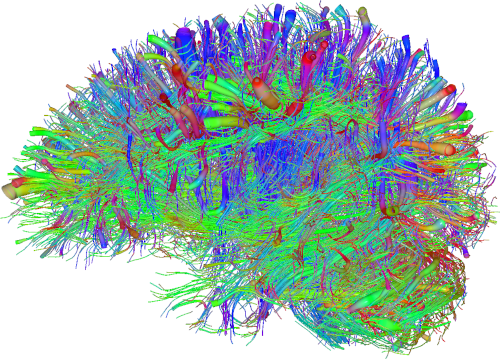
Context - Tractography from diffusion MRI is currently the only technique able to non-invasively explore the white matter architecture of the brain. It results in a tractogram, which is a set of 3D polylines (i.e., lists of ordered 3D points), usually called streamlines, which are estimates of the trajectories of large groups of nerves (axons). Indeed, current diffusion MR machines have a spatial resolution in the millimeter (mm) scale, and therefore it’s not possible to perfectly reconstruct each axon, whose diameter is typically in the micrometer (µm) scale. This means that reconstructed streamlines might approximate more than $10^5$ axons in each voxel. Nevertheless, even with such low reconstruction resolution, tractography has proven to be an invaluable tool for clinicians and researchers. It is nowadays used on a daily basis by neurosurgeons for pre-operative planning and during surgical operations. It also offers important information for studying pathological processes in neurological and psychiatric diseases and aging. When visualized, a whole-brain tractogram, namely the estimate of all the nerve streamlines in the white matter of the brain, might seem a highly intricate and complex wiring system, where it’s difficult to recognize specific bundles and a well-structured organization (see figure below). However, thanks to numerous postmortem studies, the white matter architecture of the brain has been deciphered into a set of biologically plausible pathways (also called fasciculi or tracts), that have helped understanding the functional expression of the cerebral activity. Tractogram segmentation can thus be defined as the subdivision of whole-brain tractograms into anatomically relevant and reproducible tracts with well-known structural and diffusion properties.

Goal: Explore new geometric representations, metrics and deformation models for fast, robust and reliable processing, comparison and visualization of tractograms from diffusion MRI.
Challenges: Recent tractography methods may produce whole-brain tractograms composed of millions of streamlines. This can complicate their visualization and interpretation, thus limiting the aforementioned clinical applications. Furthermore, the considerable number of streamlines can make computationally intractable processes such as segmentation (Delmonte et al., 2019), non-linear registration (Gori et al., 2016) or atlas construction (Gori et al., 2015) (Gori et al., 2017), which are important for research purposes. The presence of spurious streamlines (outliers) and the high inter-subject variability are two other challenges that can make the analysis of tractograms even more complicated. Lastly, the anatomical definition of a tract, as documented in the postmortem studies, is usually qualitative, since it is based on spatial relationships, such as “anterior to” or “close to”, that are difficult to model in a quantitative and accurate way.
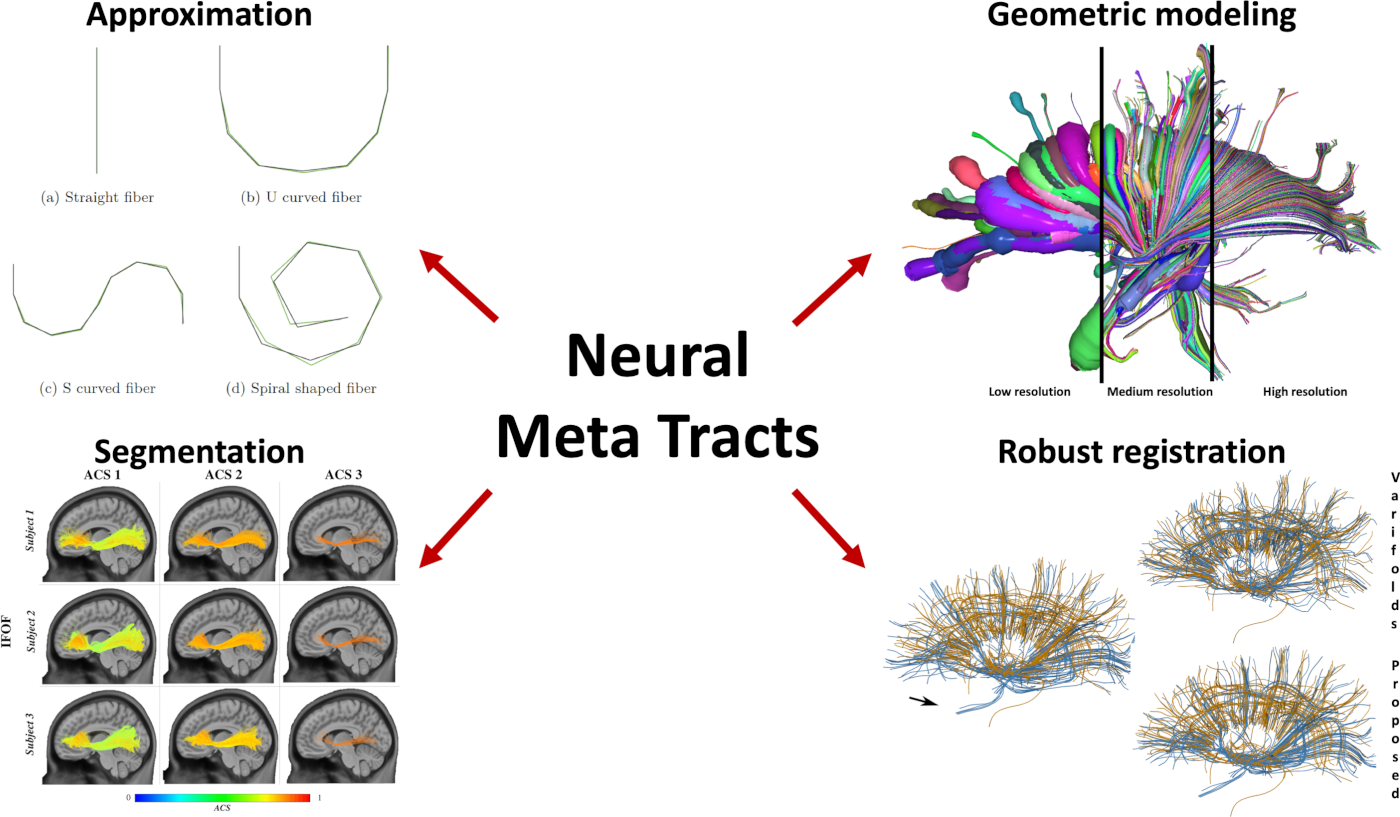
Contributions In this project, we proposed four original contributions:
- A new parsimonious and multi-resolution geometric representation for tractograms, called Neural Meta Tracts (Mercier et al., 2018)
- Qfib, a new compression algorithm well-suited for tractograms (Mercier et al., 2020)
- Two fast and scalable methods to automatically segment tractograms, one based on symbolic AI (Delmonte et al., 2019) and one on optimal transport (Feydy et al., 2019)
Press Interview published in the I’MTech, the science and technology news website of the Institut Mines-Telecom: HERE