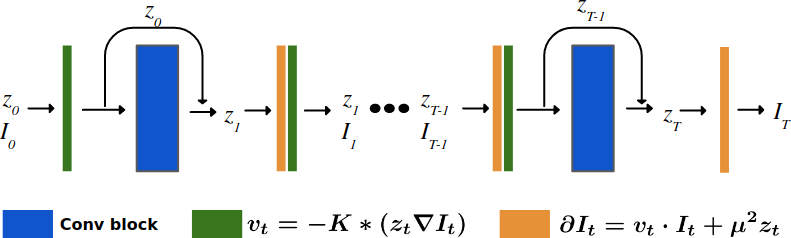Glioblastoma atlas estimation
Title : Glioblastoma atlas estimation
Project coordinator : Pietro Gori
Participants : I. Bloch (Télécom Paris), J. Glaunès (MAP5), Catherine Oppenheim (IPNP)
Institutions : Télécom Paris, Université Paris Cité and St. Anne hospital
Funding: Future & Rupture PhD Thesis grant (105 k€) + Doctoral School ED386 PhD Thesis grant (105 k€)
Period : 2019-2023

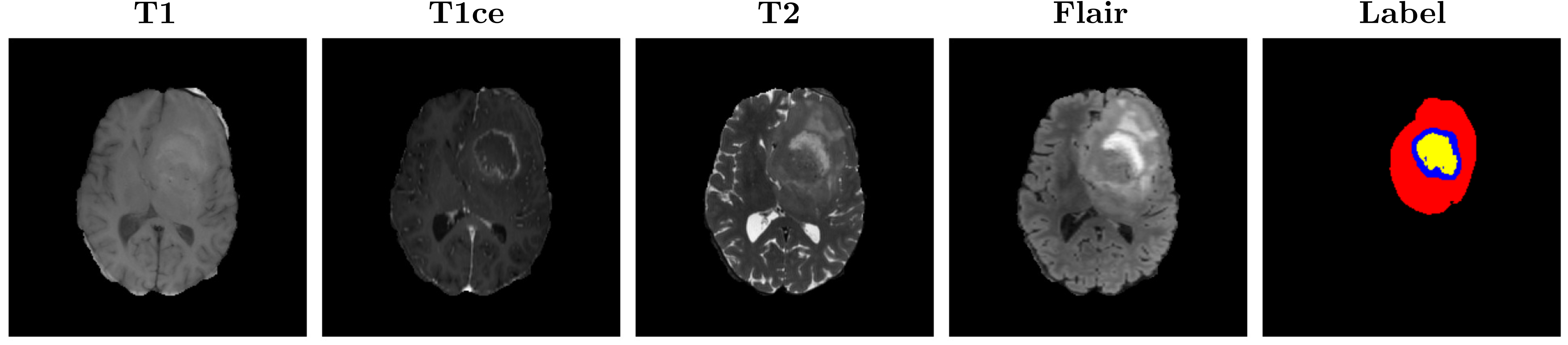
Context - Glioblastoma (GBM) is a type of aggressive brain cancer that is still considered incurable and it accounts for more than 60% of all brain tumours in adults (Roux et al., 2019). The low survival rate and negative prognosis have fostered a lot of research for a better understanding of the behavior of this kind of tumor. Clinical evidence suggests that tumor size, location, and shape could be important factors related to recurrence and seizures. The standard research protocol to detect brain tumors is Magnetic Resonance Imaging (MRI) as it constitutes a non-ionizing and non-invasive method to produce detailed images of the brain internal structures. Different MRI modalities are usually used as they provide different contrast between tissues, highlighting specific tumor parts. The commonly acquired modalities are T1, T1 contrast-enhanced (T1ce), T2, and Flair. As shown in the figure below, the contrast in each modality is different and each one highlights different tumor regions. For instance, the T1ce image shows the necrotic region and the enhancing tissue, while the Flair and T2 better reveal the edema.
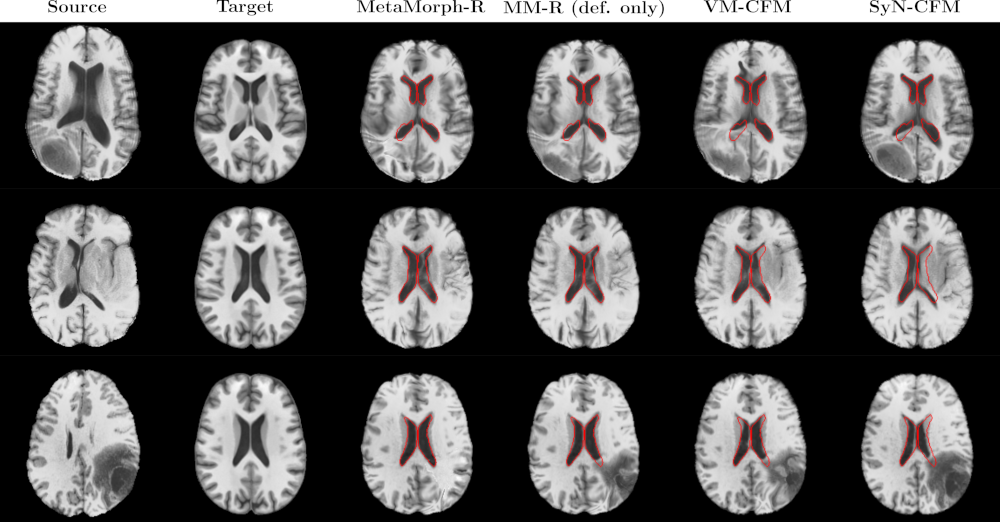
Goal: Propose a new method to estimate a 3D statistical atlas of glioblastoma using a population of MR brain images.
Challenges: In medical imaging, a statistical atlas is usually defined as an average image and a set of deformations of the average. The deformations should model the variability within the population. Most of the works in the literature focus on the morphological variability, namely the variations in shape of the anatomical structures. This analysis is relevant for modeling the healthy anatomical variability, as well as pathological variations that only concern the anatomy (e.g., atrophy in Alzheimer’s disease). Most of the works define the deformations as diffeomorphisms, which are differentiable (smooth and continuous) bijective transformation (one-to-one) with differentiable inverse. The main reason is the anatomical plausibility of the produced deformations, since they preserve the topology and spatial organization, namely no intersection, folding or shearing may occur. However, the presence of tumors induce two sources of variation that can not be taken into account by diffeomorphisms: topological and appearance changes. The first is due to the presence of tumors, since two subjects may have a different number of tumors at different locations. Appearance differences are instead due to the infiltration of the tumors causing the edema. This means that previous methods, mainly based on diffeomorphisms or splines deformations, can not be used to estimate a 3D atlas of glioblastoma.
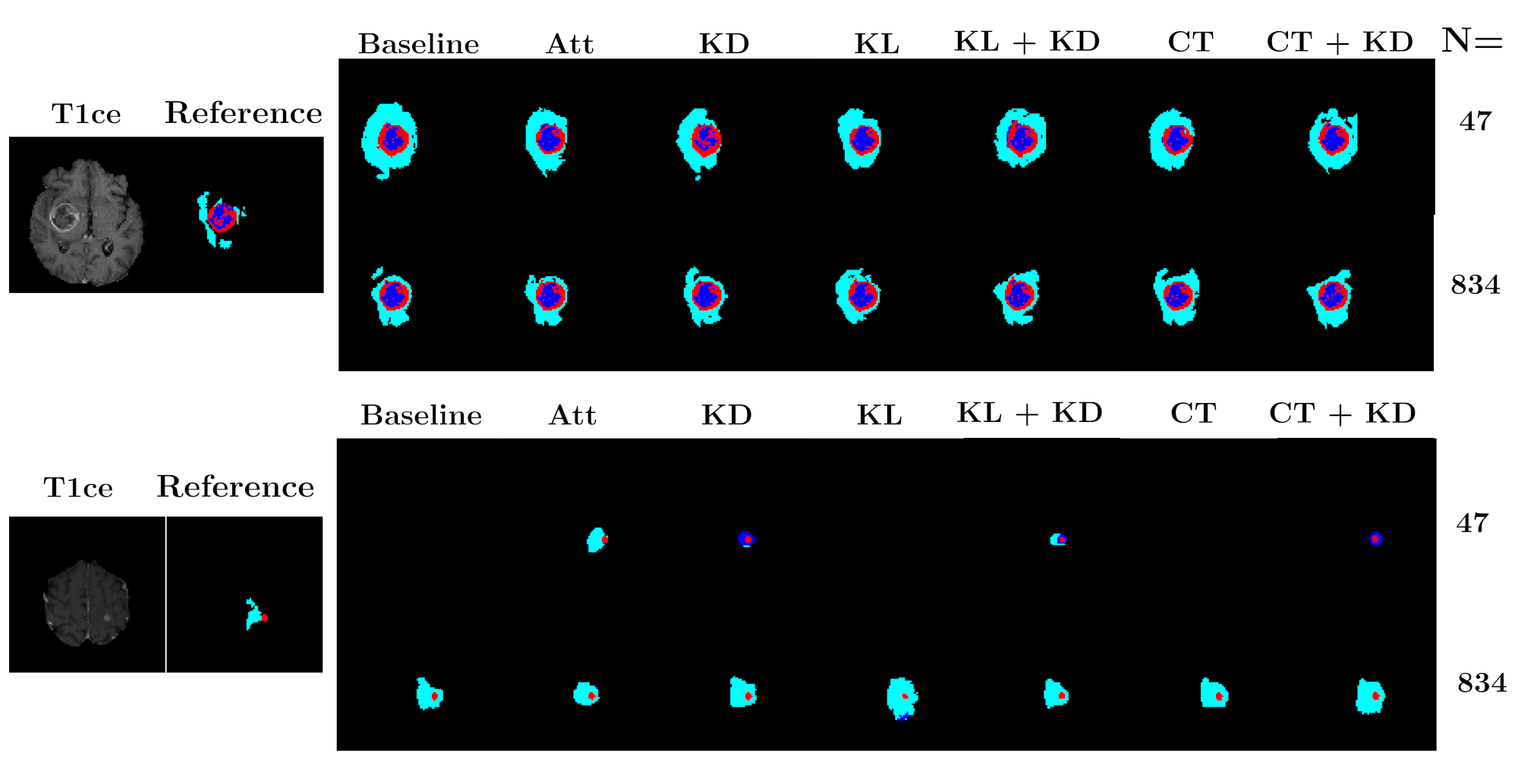
To disentangle shape and appearance variations and thus build a clinically relevant and accurate 3D atlas of glioblastoma, it is very important to correctly segment the tumor and the edema in the MR image. Multi-modal segmentation models represent the state-of-the-art technique to detect brain tumors. However, it is often difficult to obtain multiple modalities in a clinical setting due to a limited number of physicians and scanners, and to limit costs and scan time. Most of the time, only one modality is acquired.
Contributions In this project, we proposed three original contributions:
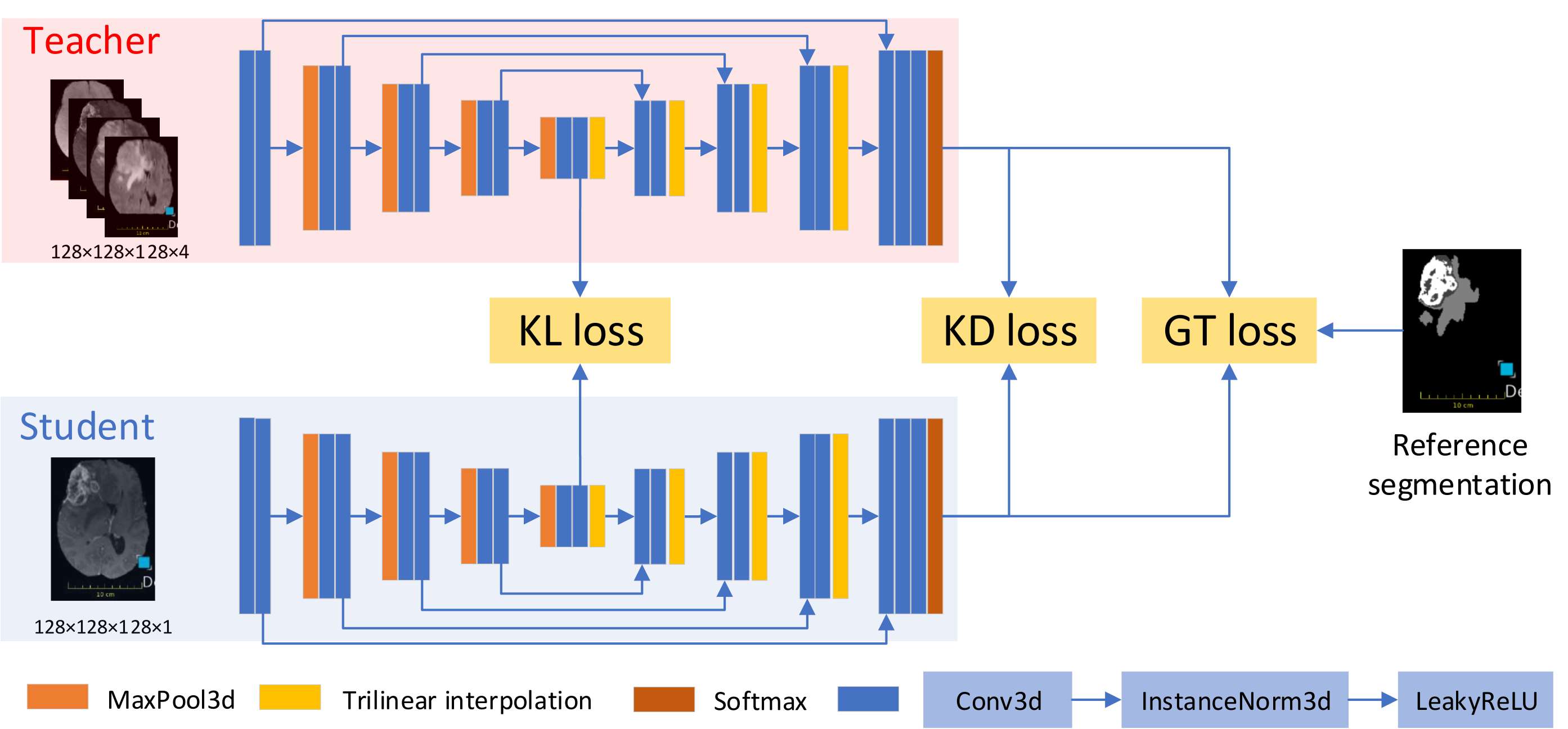
- A new framework, called KD-Net, to transfer knowledge from a multi-modal segmentation network (Teacher) to a mono-modal one (Student) (Hu et al., 2020). The student network produces a precise segmentation of all tumor areas taking as input only images from a single modality. This method can thus be used in a clinical setting, leveraging the rich datasets and computational resources available in research laboratories.
- Two implementations of the Metamorphosis image registration method based on a new semi-Lagrangian scheme (François et al., 2021) . The first uses classical numerical integration schemes (François et al., 2022) while the second employs a deep learning architecture (i.e., ResNet) (Maillard et al., 2022). Both methods leverage the KD-Net segmentation method to correctly disentangling appearance and morphological variations.